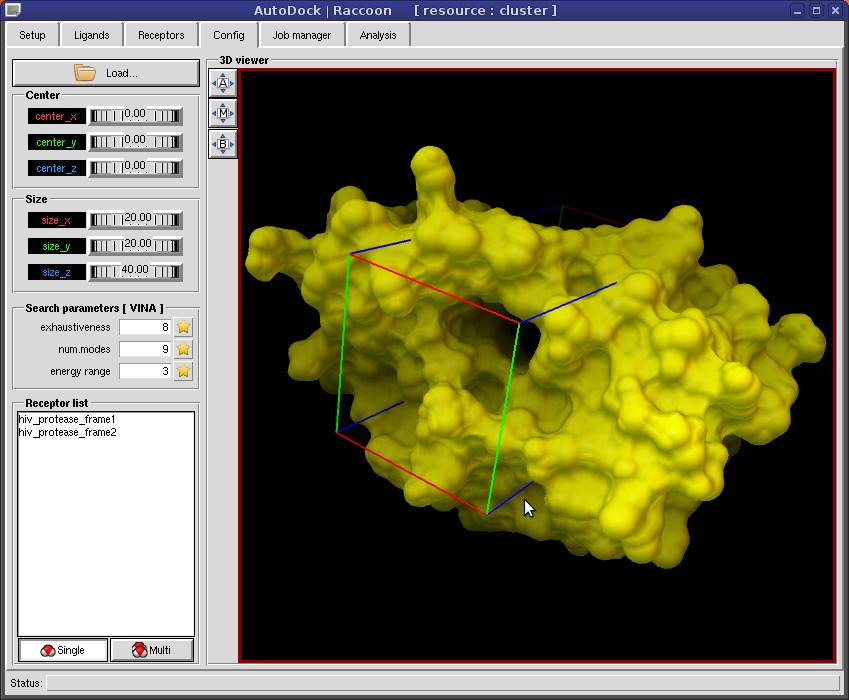
![PDF] Molecular Docking Using Chimera and Autodock Vina Software for Nonbioinformaticians | Semantic Scholar PDF] Molecular Docking Using Chimera and Autodock Vina Software for Nonbioinformaticians | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/52b25f8d3fb6c921297d2258ee9b36d05a2e33f6/17-Figure15-1.png)
PDF] Molecular Docking Using Chimera and Autodock Vina Software for Nonbioinformaticians | Semantic Scholar
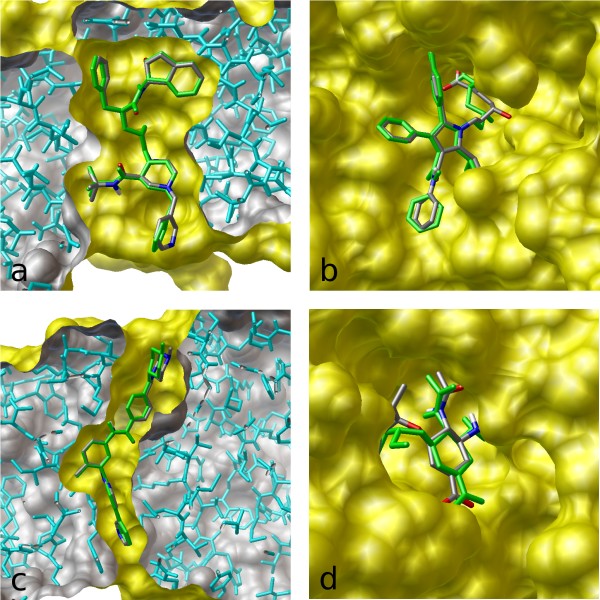
A) The output of AutoDock Vina showing the binding site residues of... | Download Scientific Diagram

AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling
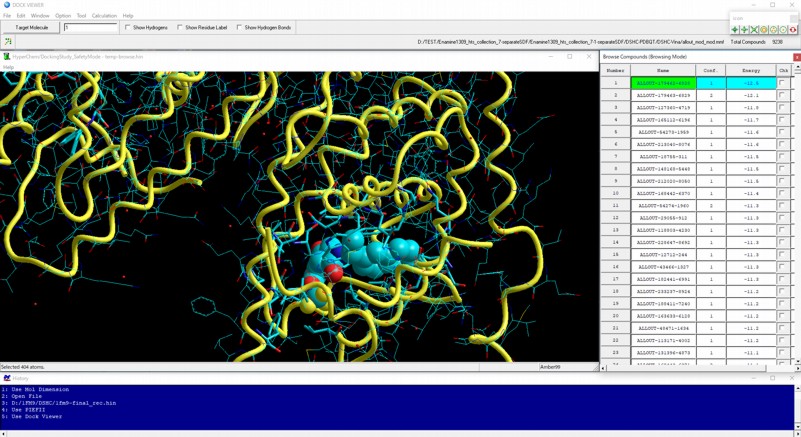
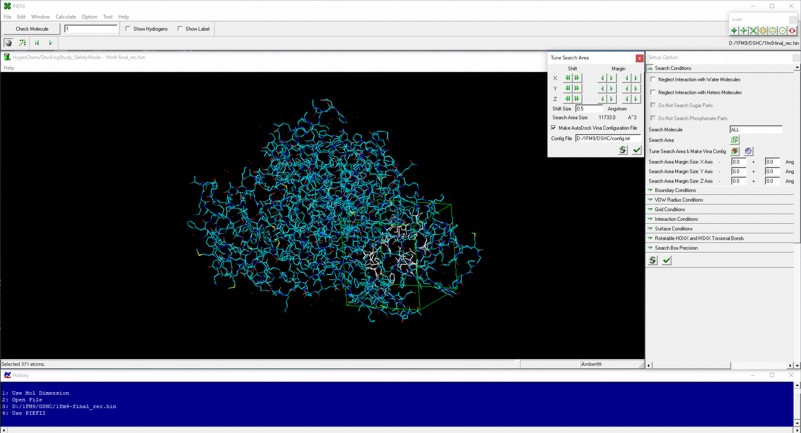
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -

SAMSON on Twitter: "AutoDock Vina in SAMSON: flexible protein-ligand docking in a few clicks http://t.co/lCyVMXt1nh" / Twitter

Computational protein-ligand docking and virtual drug screening with the AutoDock suite. - Abstract - Europe PMC

3 Autodock GUI with AutoDock-Vina by ADT. (a) Schematic representation... | Download Scientific Diagram
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -
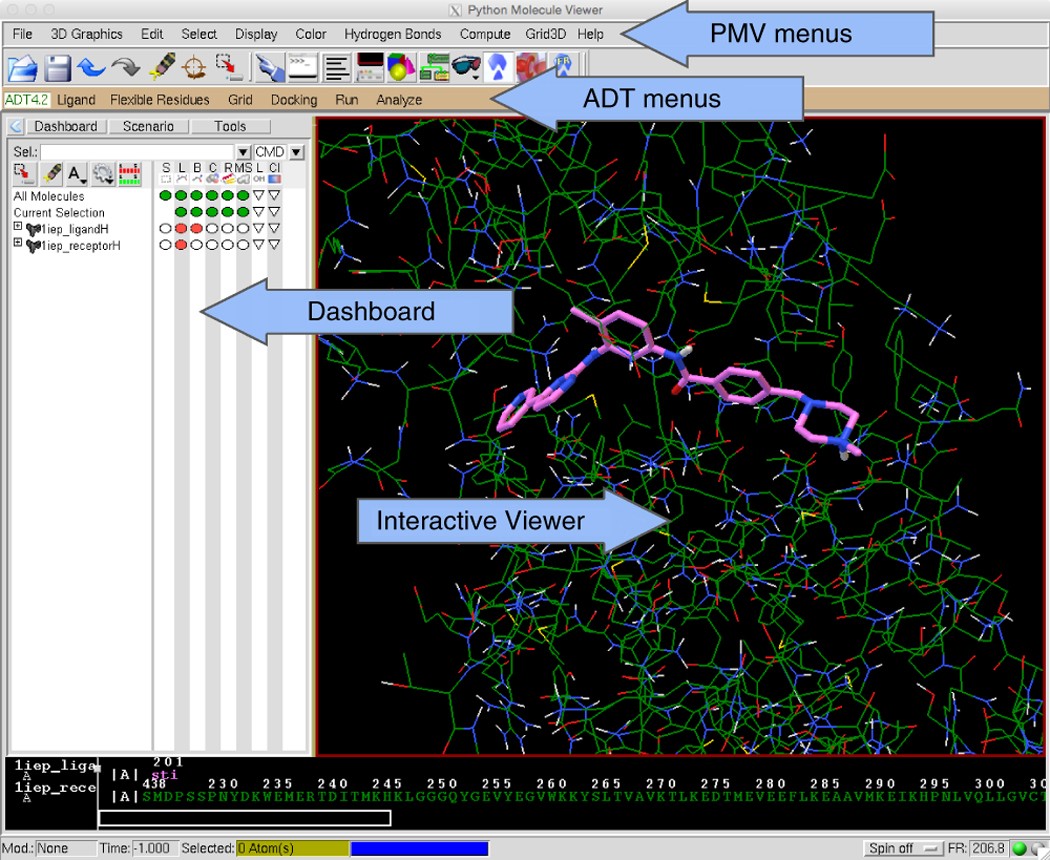
Computational protein–ligand docking and virtual drug screening with the AutoDock suite | Nature Protocols

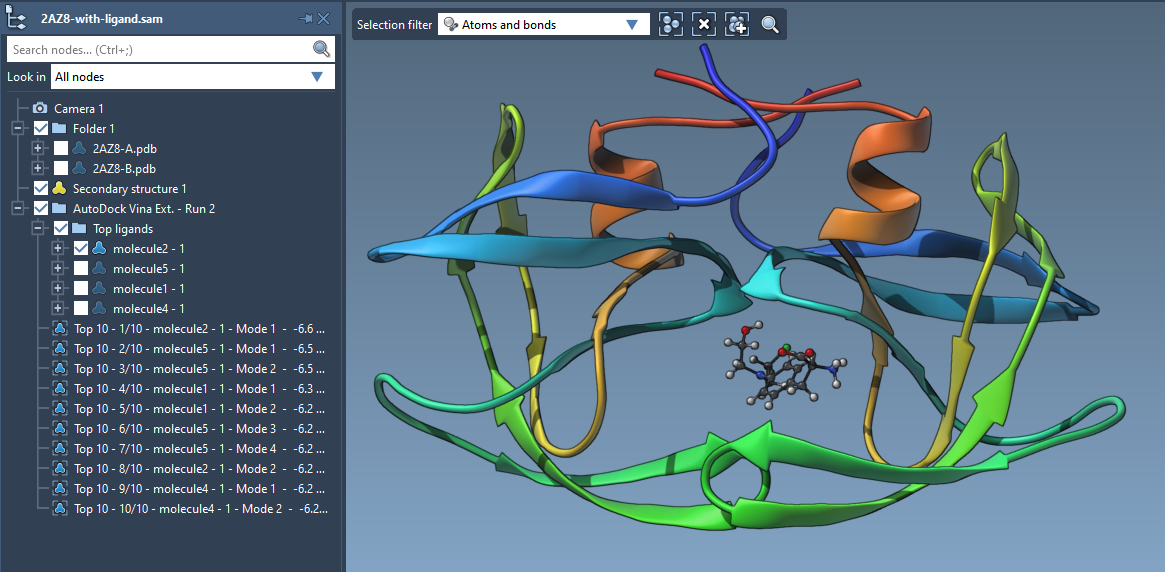
The new version of AutoDock Vina Extended is out for SAMSON 2020 R2 | The new version of AutoDock Vina Extended includes interactive analysis of docking results by linking plots to conformations.

How to install Gromacs, PyMOL, AutoDock Vina, VMD, MGLTools, Avogadro2, Open Babel in Ubuntu 20.04 - DEV Community